
Thanyada Rungrotmongkol, Associate Professor,Ph.D.
B.Sc., Kasetsart University
Ph.D., Kasetsart University
Office: Room 516/3, Science 10 Building
Phone: 662-218-5426
Fax: 662-218-5418
Email: Thanyada.r@Chula.ac.th, t.rungrotmongkol@gmail.com
Academic Experiences:
2009-2011 Researcher,Center of Innovative Nanotechnology, Chulalongkorn University, Bangkok, Thailand
2008-2009 International consultant, International Centre for Science and High Technology, United Nations Industrial Development Organization (ICS-UNIDO), Trieste, Italy
2006-2011 Postdoctoral fellow, Computational Chemistry Unit Cell, Department of Chemistry, Faculty of Sciences, Chulalongkorn University, Bangkok, Thailand
Research
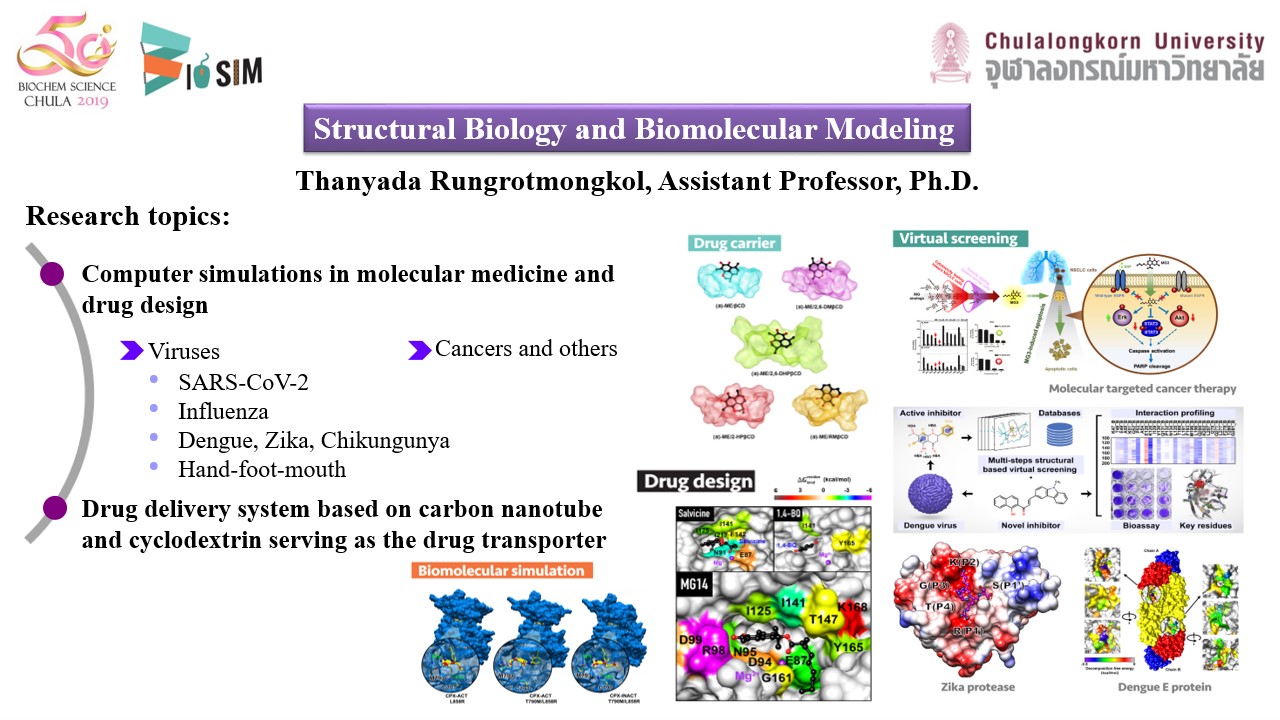
- Molecular modeling on biological systems such as influenza, chikungunya and hepatitis viruses, and cancer drug targets
- Rotational drug design against influenza neuraminidase
- Drug delivery system based on carbon nanotube andcyclodextrin serving as the drug transporter
Recent Publications
Publications
- P. Mahalapbutr, P. Chusuth, N. Kungwan, W. Chavasiri, P. Wolschann, T. Rungrotmongkol*, Molecular Recognition of Naphthoquinone-containing Compounds against Human DNA Topoisomerase IIα ATPase domain: A Molecular Modeling, Journal of Molecular Liquids, 2017, 247, 374-385.
- G. Sanober, S. Raza, T. Rungrotmongkol, S.S. Azam, The vitality of swivel domain motion in performance of Enzyme I of phosphotransferase system; A comprehensive molecular dynamic study, Journal of Molecular Liquids, 2017, 242: 1184-1198
- P. Wongpituk, B. Nutho, W. Panman, N. Kungwan, T. Rungrotmongkol*, N. Nunthaboot*, Structural dynamics and binding free energy of neral-cyclodextrins inclusion complexes: Molecular dynamics simulation, Molecular Simulation, 2017, DOI:10.1080/08927022.2017.1356458
- S. Kruawan, M. Ratanasak, R. Chanajaree, T. Rungrotmongkol, O. Saengsawang, V. Parasuk, N. Kungwan, S. Hannongbua*, Ethylene insertion in the presence of new alkoxysilane electron donors for ziegler-natta catalyzed polyethylene, Computational and Theoretical Chemistry, 2017, 1112, 10-19. DOI: 10.1016/j.comptc.2017.04.002
- S. Thompho, O. Saengsawang, T. Rungrotmongkol, N. Kungwan, S. Hannongbua*, Structure and Electronic Properties of Deformed Single-Walled Carbon Nanotubes: Quantum Calculations, Structural Chemistry, 2017, DOI:10.1007/s11224-017-0999-7
- W. Khuntawee, M. Kunaseth*, C. Rungnim, S. Intagorn, P. Wolschann, N. Kungwan, T. Rungrotmongkol*, S. Hannongbua, Comparison of Implicit and Explicit Solvation Models for Iota-Cyclodextrin Conformation Analysis from Replica Exchange Molecular Dynamics Journal of Chemical Information and Modeling, 2017, 57 (4), 778-786. DOI: 10.1021/acs.jcim.6b00595
- D. Saeloh, M. Wenzel, T. Rungrotmongkol, L.W. Hamoen, V. Tipmanee*, S.P. Voravuthikunchai*, Effects of rhodomyrtone on Gram-positive bacterial tubulin homologue. FtsZ, PeerJ, 2017, 5:e2962
- W. Karnsomwan, P. Netcharonensirisuk, T. Rungrotmongkol, W. De-Eknamkul, S. Chamni*, Synthesis, Biological Evaluation and Molecular Docking of Avicequinone C Analogues as Potential Steroid 5alpha-Reductase Inhibitors, Bulletins of the Pharmaceutical Society of Japan, 2017, 65 (3), 253-260.
- J. Phanich, T. Rungrotmongkol*, N. Kungwan, S. Hannongbua*, Role of R292K mutation in influenza H7N9 neuraminidase toward oseltamivir susceptibility: MD and MM/PB(GB)SA study, Journal of Computer-Aided Molecular Design, 2016, 30 (10), 917-926. (DOI: 10.1007/s10822-016-9981-5)
- A. Meeprasert, S. Hannongbua, N. Kungwan, T. Rungrotmongkol*, Effect of D168V Mutation in NS3/4A HCV Protease on Susceptibilities of Faldaprevir and Danoprevir, Molecular BioSystems 2016, 12 (12), 3666-3673.
- S. Thompho, T. Rungrotmongkol, O. Saengsawang, S. Hannongbua*, A Computational Study of Adsorption of Divalent Metal Ions on Graphene Oxide, Songklanakarin Journal of Science and Technology, 2016, accepted
- W. Jetsadawisut, B. Nutho, A. Meeprasert, T. Rungrotmongkol,* N. Kungwan, P. Wolschann, S. Hannongbua*, Susceptibility of inhibitors against 3C Protease of Coxsackievirus A16 and Enterovirus A71 Causing Hand, Foot and Mouth Disease: A Molecular Dynamics Study, Biophysical Chemistry, 2016, 219, 9-16.
- C. Rungnim, T. Rungrotmongkol, R. Poo-arporn*, pH-controlled doxorubicin anticancer loading and release from carbon nanotube noncovalently modified by chitosan: MD simulations, Journal of Molecular Graphics and Modelling, 2016, 70, 70-76.
- S. Tantong, O. Pringsulaka, K. Weerawanich, A. Meeprasert, T. Rungrotmongkol, R. Sarnthima, S. Roytrakul, S. Sirikantaramas. Two novel antimicrobial defensins from rice identified by genecoexpression network analyses. Peptides, 2016, 84, 7–16.
- F.N. Sabri, H. Monajemi, S.M. Zain, P.S. Wai, T. Rungrotmongkol, V.S. Lee, Molecular conformation and UV–visible absorption spectrum of emeraldine salt polyaniline as a hydrazine sensor, Integrated Ferroelectrics, 2016, 175: 1-9.
- V.T. Phuong, T. Chokbunpiam*, S. Fritzsche, T. Remsungnen, T. Rungrotmongkol, C. Chmelik, J. Caro, S. Hannongbua*, Methane in Zeolitic Imidazolate Framework ZIF-90: Adsorption and Diffusion by Molecular Dynamics and Gibbs Ensemble Monte Carlo, Microporous & Mesoporous Materials, 2016, 235, 69–77. (in press2016)
- R. Daengngern, C. Prommin; T. Rungrotmongkol, V. Promarak, P. Wolschann, N. Kungwan*, Theoretical investigation of 2-(iminomethyl)phenol in the gas phase as a prototype of ultrafast excited-state intramolecular proton transfer, Chemical Physics Letters, 2016; 657, 113-118. (DOI: 10.1016/j.cplett.2016.05.065)
- C. Rungnim, R. Chanajaree, T. Rungrotmongkol, S. Hannongbua, N. Kungwan, P. Wolschann, A. Karpfen*, V. Parasuk*, How strong is the edge effect for anti-cancer drugs adsorption over graphene cluster? Journal of Molecular Modeling, 2016, 22 (4), Article number 85. (in press2016)
- W. Karnsomwan, T. Rungrotmongkol, S. Chamni*, W. De-Eknamkul, In silico structural prediction of human steroid 5α-reductase Type II, Med. Chem. Res. 2016; 25, 1049-1056. ( DOI: 10.1007/s00044-016-1541-y)
- N. Schaduangrat, J. Phanich, T. Rungrotmongkol, H. Lerdsamran, P. Puthavathana, S. Ubol*, The significance of naturally occurring neuraminidase quasispecies of H5N1 avian influenza virus on resistance to oseltamivir: a point of concern, Journal of General Virology, 2016; 97(6):1311-23. ( DOI: 10.1099/jgv.0.000444)
- B. Nutho, A. Meeprasert, M. Chulapa, N. Kungwan, T. Rungrotmongkol*,Screening of Hepatitis C NS5B polymerase Inhibitors Containing Benzothiadiazine Core: A Steered Molecular Dynamics, Journal of Biomolecular Structure & Dynamics, 2016; 27:1-15. DOI: 10.1080/07391102.2016.1193444
- Khuntawee, W.; Rungrotmongkol, T.; Wolschann, P. ; Pongsawasdi, P.; Kungwan, N.; Okumura, H.; Hannongbua, S.. Conformation study of ε-cyclodextrin: Replica exchange molecular dynamics simulations. Carbohydrate Polymers 2016, 141, 99-105. (DOI:10.1016/j.carbpol.2015.10.018 )
- Rungnim, C.; Rungrotmongkol, T.; Kungwan, N.; Hannongbua, S.. Protein–protein interactions between SWCNT/chitosan/EGF and EGF receptor: a model of drug delivery system. Journal of Biomolecular Structure and Dynamics 2016, 34(9):1919-29. (Article in press2016) (2015 DOI : 10.1080/07391102.2015.1095114)
- J. Kicuntod, W. Khuntawee, P. Wolschann, P. Pongsawasdi, N.Kuawan, T. Rungrotmongkol* Inclusion complexation of pinostrobin with various cyclodextrin derivatives, Journal of Molecular Graphics and Modelling, 2016; 63, 91-98. (DOI: 10.1016/j.jmgm.2015.11.005)
- Phanich J.; Rungrotmongkol T.; Sindhikara D.; Phongphanphanee S.; Yoshida N.; Hirata F.; Kungwan N.; Hannongbua S.. A 3D-RISM/RISM study of the oseltamivir binding efficiency with the wild-type and resistance-associated mutant forms of the viral influenza B neuraminidase. Protein Sci. 2016, 5(1), 147-158. (2015; doi: 10.1002/pro.2718)
- W. Sangpheak, J. Kicuntod, R. Schuster, T. Rungrotmongkol, P. Wolschann, N. Kuawan, H. Viernstein, M. Mueller*, P. Pongsawasdi*, Physical properties and biological activities of hesperetin and naringenin in complex with methylated β-cyclodextrin, Beilstein Journal of Organic Chemistry, 2015; 11, 2763–2773. (in press2015)
- C. Rungnim, S. Phunpee, M. Kunaseth, S. Namuangruk, K. Rungsardthong, T. Rungrotmongkol*, U. Ruktanonchai*, Co-solvation effect on the binding mode of the α-mangostin/β-cyclodextrin inclusion complex, Beilstein Journal of Organic Chemistry, 2015; 11, 2306–2317.
- W. Khuntawee, P. Wolschann, T. Rungrotmongkol*, J. Wong-ekkabut*, S Hannongbua, Molecular dynamics simulations of the interaction of beta cyclodextrin with a lipid bilayer, Journal of Chemical Information and Modeling, 2015, 55 (9), 1894-1902.
- S. Kongkaew, P. Yotmanee, T. Rungrotmongkol, N. Kaiyawet, A. Meeprasert, T. Kaburaki, H. Noguchi, F. Takeuchi, N. Kungwan, S. Hannongbua*, Molecular Dynamics Simulation Reveals the Selective Binding of Human Leukocyte Antigen Alleles Associated with Behçet’s Disease, PLoS One, 2015; 10 (9), Article Number: e0135575
- Yotmanee, P.; Rungrotmongkol, T.; Wichapong, K.; Choi, S.B.; Wahab, H.A.; Kungwan, N.; Hannongbua, S.. Binding specificity of polypeptide substrates in NS2B/NS3pro serine protease of dengue virus type 2: A molecular dynamics Study. Journal of Molecular Graphics and Modelling 2015, 60, 24-33.
- N. Kaiyawet, R. Lonsdale, T. Rungrotmongkol, A. Mulholland*, S. Hannongbua*, High-Level QM/MM Calculations Support the Concerted Mechanism for Michael Addition and Covalent Complex Formation in Thymidylate Synthase, Journal of Chemical Theory and Computation, 2015; 11(2), 713–722
- S. Sirikataramas*, A. Meeprasert, T. Rungrotmongkol, H. Fuji, T. Hoshino, M. Yamazaki, K. Saito*, Structural insight of DNA topoisomerase I from camptothecin-producing plants revealed by molecular dynamics simulations, Phytochemistry, 2015, 113(17), 50-56.
- A. Suksuwan, L. Lomlim, T. Rungrotmongkol, T. Nakpheng, F.L. Dickert, S. Roongnapa*, The Composite Nanomaterials containing (R)-Thalidomide-Molecularly Imprinted Polymers as a Recognition System for Enantioselective-Controlled Release and Targeted Drug Delivery, Journal of Applied Polymer Science, 2015; 132(18), 41930(1)-(21)
- Ratanasak, M.; Rungrotmongkol, T.; Saengsawang, O.; Hannongbua, S.; Parasuk, V.. Towards the design of new electron donors for Ziegler-Natta catalyzed propylene polymerization using QSPR modeling. Polymer (United Kingdom) 2015, 56, 340-345.
- Rungrotmongkol, T.; Mulholland, A.J.; Hannongbua, S.. QM/MM simulations indicate that Asp185 is the likely catalytic base in the enzymatic reaction of HIV-1 reverse transcriptase. MedChemComm 2014, 5 (5), 593-596.
- B. Nutho, W. Khuntawee, C. Rungnim, P. Pongsawasdi, P. Wolschann, A. Karpfen, T. Rungrotmongkol*, Binding mode and free energy prediction of fisetin/β-cyclodextrin inclusion complex, Beilstein Journal of Organic Chemistry, 2014; 10, 2789–2799.
- Sangpheak, W.; Khuntawee, W.; Wolschann, P.; Pongsawasdi, P.; Rungrotmongkol, T.. Enhanced stability of a naringenin/2,6-dimethyl beta-cyclodextrin inclusion complex: Molecular dynamics and free energy calculations based on MM- and QM-PBSA/GBSA. JOURNAL OF MOLECULAR GRAPHICS & MODELLING 2014, 50, 10-15.
- Meeprasert, A.; Hannongbua, S.; Rungrotmongkol, T.. Key Binding and Susceptibility of NS3/4A Serine Protease Inhibitors against Hepatitis C Virus. JOURNAL OF CHEMICAL INFORMATION AND MODELING 2014, 54 (4), 1208-1217.
- Maitarad, P.; Han, J.; Zhang, D. ; Shi, L.; Namuangruk, S.; Rungrotmongkol, T.. Structure-activity relationships of NiO on CeO2 nanorods for the selective catalytic reduction of NO with NH3: Experimental and DFT studies. Journal of Physical Chemistry C 2014, 118 (18), 9612-9620.
- Meeprasert A.; Rungrotmongkol T.; Li M. S.; Hannongbua S.. In silico screening for Potent Inhibitors against the NS3/4A Protease of Hepatitis C Virus. Current Pharmaceutical Design 2014, 20 (21), 3465-3477.
- Udommaneethanak T.; Rungrotmongkol T.; Frecer V.; Seneci P.; Stanislav M.; Bren U.. Drugs Against Avian Influenza A Virus : Design of Novel Sulfonate Inhibitors of Neuraminidas N1. Current Pharmaceutical Design2014, 20 (21), 3478-3487.
- N. Nunthaboot, T. Rungrotmongkol, O. Aruksakunwong, S. Hannongbua*, Effects of protonation state of catalytic residues and ligands upon binding and recognition in targeted proteins of HIV-1 and influenza viruses, Current Pharmaceutical Design 2013, 19(42), 4276-4290.
- Kaiyawet, N.; Rungrotmongkol, T.; Hannongbua, S. Effect of halogen substitutions on dUMP to stability of thymidylate synthase/dUMP/mTHF ternary complex using molecular dynamics simulation. Journal of Chemical Information and Modeling 2013, 53(6), 1315-1323.
- Gao R.; Zhang D.; Maitarad P.; Shi L., Rungrotmongkol T.; Li H.; Zhang J. and Cao W. Morphology-Dependent Properties of MnOx/ZrO2−CeO2 Nanostructures for the Selective Catalytic Reduction of NO with NH3, J. Phys. Chem C 2013, 117(20), 10502-10511.
- Maitarad P.; Zhang D.; Gao R.; Shi L.; Li H.; Huang L.; Rungrotmongkol T.; and Zhang J. Combination of Experimental and Theoretical Investigations of MnOx/Ce0.9Zr0.1O2 Nanorods for Selective Catalytic Reduction of NO with Ammonia. J. Phys. Chem. C 2013, 117, 9999−10006.
- Kaiyawet, N.; Rungrotmongkol, T.; and Hannongbua , S. Probable polybasic residues inserted into the cleavage site of the highly pathogenic avian influenza A/H5N1 hemagglutinin: speculation of the next outbreak in humans. International Journal of Quantum Chemistry 2013, 113(4), 569-573.
- Rungnim, C .; Rungrotmongkol, T.; Hannongbua, S.; Okumura, H. Replica exchange molecular dynamics simulation of chitosan for drug delivery system based on carbon nanotube. Journal of molecular graphics and modeling 2013, 39, 183-192.
- C . Rungnim, U, Arsawang, T. Rungrotmongkol*, S Hannongbua, Molecular dynamics properties of varying amounts of the anticancer drug gemcitabine inside an open-ended single-walled carbon nanotube, Chemical Physics Letter 2012; 550: 99-103.
- A. Meeprasert, W. Khuntawee, K. Kamlungsua, N. Nunthaboot, T. Rungrotmongkol*, S. Hannongbua, Binding pattern of the long acting neuraminidase inhibitor laninamivir toward influenza A subtypes H5N1 and pandemic H1N1, Journal of molecular graphics and modeling 2012;38: 148-154.
- J. Kongkamnerd, L. Cappelletti, A. Prandi, P. Seneci*, T. Rungrotmongkol, N. Jongaroonngamsang, P. Rojsitthisak, V. Frecer*, A. Milani, G. Cattoli, C. Terregino, I. Capua, L. Beneduce, A. Gallotta, P. Pengo, G. Fassina, S. Miertus, W. De-Eknamkul*, Synthesis and in vitro study of novel neuraminidase inhibitors against avian influenza virus, Bioorganic & Medicinal Chemistry 2012;20: 2152–2157.
- Vongachariya, A.; Iamsamai, C.; Saengsawang, O.; Rungrotmongkol, T.; Dubas, S.; Parasuk, V.; and Hannongbua, S.The surface curvature effect of single-walled carbon nanotube on its cation-pi interaction with monovalent cations, Journal of Computational and Theoretical Nanosciencs 2012, 9, 2107-2112.
- P. Kongsune, T. Rungrotmongkol, N. Nunthaboot, P. Yotmanee, P. Sompornpisut, Y. Poovorawan, P. Wolschann and S. Hannongbua*, Molecular insights into the binding affinity and specificity of high pathogenic H5N1 hemagglutinin cleavage loop toward proprotein convertase furin, Monatshefte fur Chemie – Chemical Monthly2012;143:853–860.
- Khuntawee W.; Rungrotmongkol T.; Hannongbua., Molecular Dynamic Behavior and Binding Affinity of Flavonoid Analogues to the Cyclin Dependent Kinase 6/cyclin D Complex, Journal of Chemical Information and Modeling 2012, 52, 76-83.
- T. Rungrotmongkola, P. Yotmaneea, N. Nunthaboota and S. Hannongbua *, Computational studies of influenza A virus at three important targets: hemagglutinin, neuraminidase and M2 protein, Current Pharmaceutical Design2011;17(17):1720-1739.
- T. Rungrotmongkol, U. Arsawang, C. Iamsamai, A. Vongachariya, S. Dubas, U. Ruktanonchai, A. Soottitantawat and S. Hannongbua*, Increase in dispersion and solubility of carbon nanotubes noncovalently modified by chitosan-polysaccharide biopolymer: MD simulations, Chemical Physics Letter 2011;507(1-3):134–137.
- U. Arsawang, O. Saengsawang, T. Rungrotmongkol, P. Sornmee, K. Wittayanarakul, T. Remsungnen and S. Hannongbua*, How do carbon nanotubes serve as carriers for gemcitabine transport in drug delivery system? Journal of molecular graphics and modelling 2011;29(5):591–596.
- P. Sornmee, T. Rungrotmongkol, O. Saengsawang, U. Arsawang, T. Remsungnen and S. Hannongbua*, Understanding of molecular properties of the doxorubicin anticancer drug filling inside and wrapping outside the single-walled carbon nanotube Journal of Computational and Theoretical Nanosciencs 2011;8(8):1385-1391.
- T. Rungrotmongkol*, N. Nunthaboot, M. Malaisree, N. Kaiyawet, P. Intharathep, A. Meepraset and S. Hannongbua, Molecular Insight into the Specific Binding of ADP-ribose to the nsP3 Macro Domains of Chikungunya and Venezuelan Equine Encephalitis Viruses: Molecular Dynamics Simulations and Free Energy Calculations Journal of molecular graphics and modelling 2010;29(3):347–353.
- N. Nunthaboot, T. Rungrotmongkol, M. Malaisree, N. Kaiyawet, P. Decha, P. Sompornpisut, and S. Hannongbua*, Evolution of the human-receptor binding affinity of influenza A (H1N1) 2009 pandemic hemagglutinin from 1918- and 1930-H1N1 influenza viruses, Journal of Chemical Information and Modeling 2010;50(8):1410-1417.
- S. Phongphanphanee, T. Rungrotmongkol, N. Yoshida, S. Hannongbua, F. Hirata*, Proton transport through the influenza A M2 channel: 3D-RISM study, Journal of American Chemical Society 2010; 132(28):9782-9788.
- P. Intharathep, T. Rungrotmongkol, P. Decha, N. Nunthaboot, N. Kaiyawet, T. Kerdcharoen, P. Sompornpisut, and S. Hannongbua*, Evaluating how rimantadines control the proton gating of the Influenza A M2-proton port via allosteric binding outside of the M2-channel: MD simulations, Journal of Enzyme Inhibition and Medicinal Chemistry 2011; 26(2):162-168.
- N. Nunthaboot, T. Rungrotmongkol, M. Malaisree, P. Decha, N. Kaiyawet, P. Intharathep, P. Sompornpisut, Y. Poovorawan and S. Hannongbua*, Molecular insights into human receptor binding to 2009 H1N1 influenza A hemagglutinin, Monatshefte fur Chemie – Chemical Monthly 2010; 141(7):801-807.
- T. Rungrotmongkola, M. Malaisreea, N. Nunthaboot, P. Sompornpisut, and S. Hannongbua*, Molecular prediction of oseltamivir efficiency against probable influenza A (H1N1-2009) mutants: Molecular modelling approach, Amino Acids 2010; 39(2):393-398.
- T. Rungrotmongkol, T. Udommaneethanakit, V. Frecer, and S. Miertus, Combinatorial design of avian influenza neuraminidase inhibitors containing pyrrolidine core with a reduced susceptibility to viral drug resistance, Combinatorial Chemistry & High Throughput Screening 2010; 13(3):268-77.
- T. Udommaneethanakit, T. Rungrotmongkol, U. Bren, V. Frecer and S. Miertus*, Dynamic Behavior of Avian Influenza A Virus Neuraminidase Subtype H5N1 in Complex with Oseltamivir, Zanamivir, Peramivir, and their Phosphonate Analogues, Journal of Chemical Information and Modeling 2009; 49:2323–2332.
- T. Rungrotmongkol, T. Udommaneethanakit, M. Malaisree, N. Nunthaboot, P. Intharathep, P. Sompornpisut and S. Hannongbua*, How does each substituent functional group of oseltamivir lose its activity against virulent H5N1 influenza mutants? Biophysical Chemistry 2009; 145(1):29–36.
- T. Rungrotmongkol, P. Intharathep, M. Malaisree, N. Nunthaboot, N. Kaiyawet, P. Sompornpisut, S. Payungporn, Y. Poovorawan, and S. Hannongbua*, Susceptibility of antiviral drugs against 2009 influenza A (H1N1) virus.Biochemical and Biophysical Research Communications 2009; 385(3):390-4.
- T. Rungrotmongkol, V. Frecer, W. De-Eknamkul, S. Hannongbua, and S. Miertus*, Design and in silico screening of combinatorial library of oseltamivir analogs inhibiting neuraminidase of avian influenza virus H5N1. Antiviral Research 2009; 82(1):51-8.
- T. Rungrotmongkol, M. Malaisree, T. Udommaneethanakit and S. Hannongbua*, Comment on “Another look at the molecular mechanism of the resistance of H5N1 influenza A virus neuraminidase (NA) to oseltamivir (OTV)” Biophysical Chemistry 2009; 141: 131-132.
- C. Laohpongspaisan, T. Rungrotmongkol, P. Intharathep, M. Malaisree, P. Decha, O. Aruksakunwong, P. Sompornpisut, S. Hannongbua*, Why amantadine loses its function in influenza M2 mutants: MD simulations. Journal of Chemical Information and Modeling 2009; 49(4):847-52.
- T. Rungrotmongkol, P. Decha, P. Sompornpisut, M. Malaisree, P. Intharathep, N. Nunthaboot, T. Udommaneethanakit, O. Aruksakunwong, and S. Hannongbua*, Combined QM/MM mechanistic study of the acylation process in furin complexed with the H5N1 avian influenza virus hemagglutinin’s cleavage site. Proteins: Structure, Function, and Bioinformatics 2009; 76(1): 62-71.
- M. Malaisreea, T. Rungrotmongkola, N. Nunthaboot, O. Aruksakunwong, P. Intharathep, P. Decha, P. Sompornpisut and S. Hannongbua*, Source of Oseltamivir Resistance in Avian Influenza H5N1 Virus with the H274Y Mutation. Amino Acids 2009; 37(4):725–732.
- T. Rungrotmongkol, P. Decha, M. Malaisree, P. Sompornpisut, and S. Hannongbua*, Comment on ‘‘Cleavage mechanism of the H5N1 hemagglutinin by trypsin and furin” [Amino Acids 2008, January 31, Doi: 10.1007=s00726-007-0611-3], Amino Acids 2008; 35: 511-512.
- V. Nukoolkarn, S. Saen-oon, T. Rungrotmongkol, S. Hannongbua, K. Ingkaninan, K. Suwanborirux*, Petrosamine, a potent anticholinesterase pyridoacridine alkaloid from a Thai marine sponge Petrosia n. sp., Bioorganic & Medicinal Chemistry 2008; 16(13): 6560-6567.
- P. Intharathep, C. Laohpongspaisan, T. Rungrotmongkol, A. Loisruangsin, M. Malaisree, P. Decha, O. Aruksakunwong, K. Chuenpennit, N. Kaiyawet, P. Sompornpisut, S. Pianwanit, and S. Hannongbua*, How amantadine and rimantadine inhibit proton transport in the M2 protein channel, Journal of molecular graphics and modelling 2008; 27(3): 342-348.
- P. Decha, T. Rungrotmongkol, P. Intharathep, M. Malaisree, O. Aruksakunwong, C. Laohpongspaisan, V. Parasuk, P.Sompornpisut, S. Pianwanit, S. Kokpol and S. Hannongbua*, Source of high pathogenicity of an avian influenza virus H5N1: Why H5 is better cleaved by furin, Biophysical Journal 2008; 95(1): 128-134.
- M. Malaisree, T. Rungrotmongkol, P. Decha, P. Intharathep, O. Aruksakunwong, and S. Hannongbua*, Understanding of known drug-target interactions in the catalytic pocket of neuraminidase subtype N1, Proteins: Structure, Function, and Bioinformatics 2008; 71(4): 1908-1918.
- T. Rungrotmongkol, A. J. Mulholland and S. Hannongbua*, Active site dynamics and combined quantum mechanics/molecular mechanics (QM/MM) modelling of a HIV-1 reverse transcriptase/DNA/dTTP complex, Journal of molecular graphics and modelling 2007; 26(1): 1-13.
- T. Rungrotmongkol, S. Hannongbua* and A. J. Mulholland, Mechanistic study of HIV-1 reverse transcriptase at the active site based on QM/MM method, Journal of Theoretical and Computational Chemistry 2004; 3(4): 491-500.

